Python 机器学习
层次聚类简介
层次聚类是另一种无监督学习算法,用于将具有相似特征的未标记数据点分组。层次聚类算法分为以下两类:
- 凝聚式层次算法:在凝聚式层次算法中,每个数据点被视为一个单独的簇,然后逐步合并或聚集(自下而上方法)成对的簇。簇的层次结构表示为树状图或树形结构。
- 分裂式层次算法:另一方面,在分裂式层次算法中,所有数据点被视为一个大的簇,聚类过程涉及将一个大的簇划分为各种小簇(自上而下方法)。
执行凝聚式层次聚类的步骤
我们将解释最常用且重要的层次聚类,即凝聚式聚类。执行该聚类的步骤如下:
步骤 1:将每个数据点视为一个单独的簇。因此,开始时我们会有 K 个簇。数据点的数量开始时也将是 K。
步骤 2:现在,在此步骤中,我们需要通过连接两个最近的数据点来形成一个大簇。这将导致总共有 K-1 个簇。
步骤 3:现在,为了形成更多的簇,我们需要连接两个最近的簇。这将导致总共有 K-2 个簇。
步骤 4:现在,为了形成一个大的簇,重复前面三个步骤,直到 K 变为 0,即没有更多数据点可供连接。
步骤 5:最后,在形成一个单一的大簇后,将根据问题使用树状图将其划分为多个簇。
树状图在凝聚式层次聚类中的作用
正如我们在最后一步讨论的那样,一旦形成大簇,树状图的作用就开始了。树状图将用于根据我们的问题将簇分割成多个相关数据点的簇。这可以通过以下示例来理解:
示例 1
为了理解,让我们首先导入所需的库,如下所示:
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
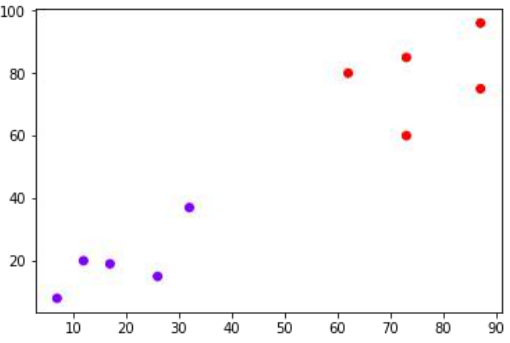
接下来,我们将绘制为此示例选取的数据点:
X = np.array([[7,8],[12,20],[17,19],[26,15],[32,37],[87,75],[73,85],
[62,80],[73,60],[87,96],])
labels = range(1, 11)
plt.figure(figsize=(10, 7))
plt.subplots_adjust(bottom=0.1)
plt.scatter(X[:,0],X[:,1], label='True Position')
for label, x, y in zip(labels, X[:, 0], X[:, 1]):
plt.annotate(label,xy=(x, y), xytext=(-3, 3),textcoords='offset points',
ha='right', va='bottom')
plt.show()
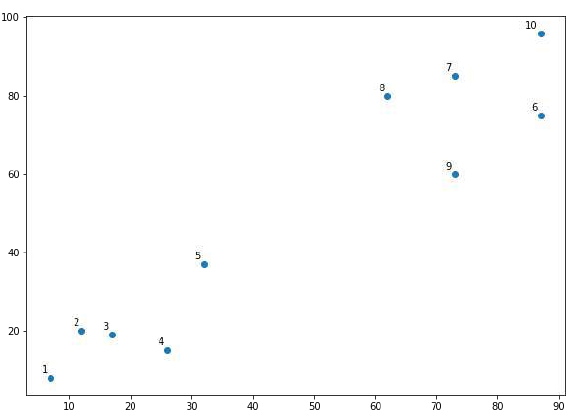
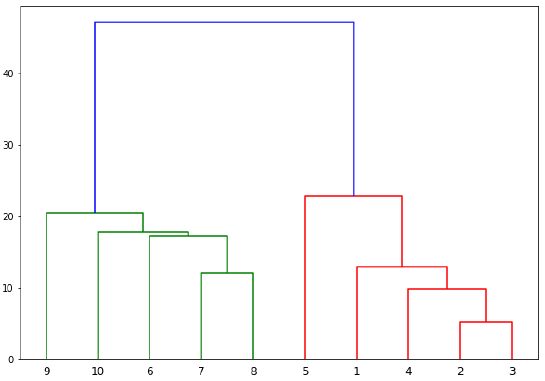
从上图中,很容易看出我们的数据点中有两个簇,但在真实世界的数据中,可能存在成千上万个簇。接下来,我们将使用 Scipy 库绘制数据点的树状图:
from scipy.cluster.hierarchy import dendrogram, linkage
from matplotlib import pyplot as plt
linked = linkage(X, 'single') # 使用 'single' 连接方法
labelList = range(1, 11)
plt.figure(figsize=(10, 7))
dendrogram(linked, orientation='top',labels=labelList,
distance_sort='descending',show_leaf_counts=True)
plt.show()
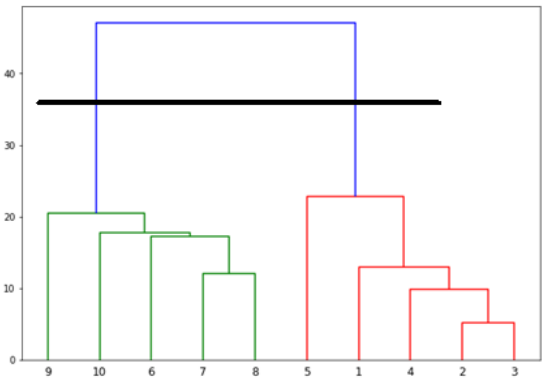
现在,一旦形成了大簇,就选择最长的垂直距离。然后穿过它绘制一条垂直线,如下图所示。由于水平线在两点处穿过蓝线,因此簇的数量将是两个。
接下来,我们需要导入用于聚类的类并调用其 fit_predict 方法来预测簇。我们正在导入 sklearn.cluster 库的 AgglomerativeClustering 类:
from sklearn.cluster import AgglomerativeClustering
cluster = AgglomerativeClustering(n_clusters=2, affinity='euclidean',
linkage='ward') # affinity 在 sklearn 1.2+ 中已弃用,会被忽略,或可以使用 metric 参数
cluster.fit_predict(X)
接下来,使用以下代码绘制簇:
plt.scatter(X[:,0],X[:,1], c=cluster.labels_, cmap='rainbow')
plt.show()
上述图表显示了我们数据点中的两个簇。
示例 2
正如我们从上面讨论的简单示例中理解了树状图的概念一样,让我们转到另一个示例,在该示例中,我们将使用层次聚类在 Pima Indian Diabetes 数据集中创建数据点的簇:
import matplotlib.pyplot as plt
import pandas as pd
%matplotlib inline
import numpy as np
from pandas import read_csv
path = r"C:\pima-indians-diabetes.csv"
headernames = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age',
'class']
data = read_csv(path, names=headernames)
array = data.values
X = array[:,0:8] # 所有行,前8列作为特征
Y = array[:,8] # 所有行,第9列作为目标(类别)
print(data.shape)
print(data.head())
输出:
(768, 9)
preg plas pres skin test mass pedi age class
0 6 148 72 35 0 33.6 0.627 50 1
1 1 85 66 29 0 26.6 0.351 31 0
2 8 183 64 0 0 23.3 0.672 32 1
3 1 89 66 23 94 28.1 0.167 21 0
4 0 137 40 35 168 43.1 2.288 33 1
选择用于聚类的数据子集(示例中使用了 'skin' 和 'test' 列):
patient_data = data.iloc[:, 3:5].values # 提取第3列和第4列作为患者数据
import scipy.cluster.hierarchy as shc
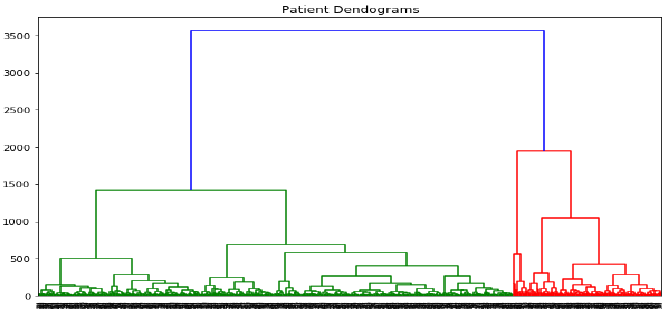
plt.figure(figsize=(10, 7))
plt.title("Patient Dendograms")
dend = shc.dendrogram(shc.linkage(data, method='ward')) # 对整个数据集进行 linkage
plt.show()
from sklearn.cluster import AgglomerativeClustering
# affinity 在 sklearn 1.2+ 中已弃用,会被忽略。如果需要指定距离度量,请使用 metric 参数。
cluster = AgglomerativeClustering(n_clusters=4, affinity='euclidean',
linkage='ward')
cluster.fit_predict(patient_data) # 对 patient_data 进行聚类
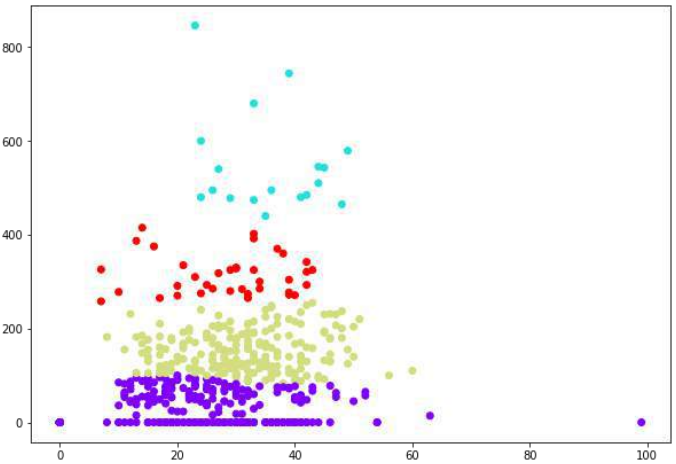
plt.figure(figsize=(10, 7))
plt.scatter(patient_data[:,0], patient_data[:,1], c=cluster.labels_,
cmap='rainbow')
plt.show()
20. Clustering Algorithms – Hierarchical
Machine Learning Clustering
with Python
Introduction to Hierarchical Clustering
Hierarchical clustering is another unsupervised learning algorithm that is used to group
together the unlabeled data points having similar characteristics. Hierarchical clustering
algorithms falls into following two categories:
Agglomerative hierarchical algorithms: In agglomerative hierarchical algorithms,
each data point is treated as a single cluster and then successively merge or agglomerate
(bottom-up approach) the pairs of clusters. The hierarchy of the clusters is represented
as a dendrogram or tree structure.
Divisive hierarchical algorithms: On the other hand, in divisive hierarchical algorithms,
all the data points are treated as one big cluster and the process of clustering involves
dividing (Top-down approach) the one big cluster into various small clusters.
Steps to Perform Agglomerative Hierarchical Clustering
We are going to explain the most used and important Hierarchical clustering i.e.
agglomerative. The steps to perform the same is as follows:
Step1: Treat each data point as single cluster. Hence, we will be having, say K clusters at
start. The number of data points will also be K at start.
Step2: Now, in this step we need to form a big cluster by joining two closet datapoints.
This will result in total of K-1 clusters.
Step3: Now, to form more clusters we need to join two closet clusters. This will result in
total of K-2 clusters.
Step4: Now, to form one big cluster repeat the above three steps until K would become 0
i.e. no more data points left to join.
Step5: At last, after making one single big cluster, dendrograms will be used to divide
into multiple clusters depending upon the problem.
Role of Dendrograms in Agglomerative Hierarchical Clustering
As we discussed in the last step, the role of dendrogram starts once the big cluster is
formed. Dendrogram will be used to split the clusters into multiple cluster of related data
points depending upon our problem. It can be understood with the help of following
example:
Example1
To understand, let us start with importing the required libraries as follows:
%matplotlib inline
124
Machine Learning with Python
import matplotlib.pyplot as plt
import numpy as np
Next, we will be plotting the datapoints we have taken for this example:
X = np.array([[7,8],[12,20],[17,19],[26,15],[32,37],[87,75],[73,85],
[62,80],[73,60],[87,96],])
labels = range(1, 11)
plt.figure(figsize=(10, 7))
plt.subplots_adjust(bottom=0.1)
plt.scatter(X[:,0],X[:,1], label='True Position')
for label, x, y in zip(labels, X[:, 0], X[:, 1]):
plt.annotate(label,xy=(x, y), xytext=(-3, 3),textcoords='offset points',
ha='right', va='bottom')
plt.show()
From the above diagram, it is very easy to see that we have two clusters in out datapoints
but in the real world data, there can be thousands of clusters. Next, we will be plotting the
dendrograms of our datapoints by using Scipy library:
from scipy.cluster.hierarchy import dendrogram, linkage
from matplotlib import pyplot as plt
linked = linkage(X, 'single')
labelList = range(1, 11)
plt.figure(figsize=(10, 7))
125
Machine Learning with Python
dendrogram(linked, orientation='top',labels=labelList,
distance_sort='descending',show_leaf_counts=True)
plt.show()
Now, once the big cluster is formed, the longest vertical distance is selected. A vertical
line is then drawn through it as shown in the following diagram. As the horizontal line
crosses the blue line at two points, the number of clusters would be two.
Next, we need to import the class for clustering and call its fit_predict method to predict
the cluster. We are importing AgglomerativeClustering class of sklearn.cluster
library:
126
Machine Learning with Python
from sklearn.cluster import AgglomerativeClustering
cluster = AgglomerativeClustering(n_clusters=2, affinity='euclidean',
linkage='ward')
cluster.fit_predict(X)
Next, plot the cluster with the help of following code:
plt.scatter(X[:,0],X[:,1], c=cluster.labels_, cmap='rainbow')
The above diagram shows the two clusters from our datapoints.
Example2
As we understood the concept of dendrograms from the simple example discussed above,
let us move to another example in which we are creating clusters of the data point in Pima
Indian Diabetes Dataset by using hierarchical clustering:
import matplotlib.pyplot as plt
import pandas as pd
%matplotlib inline
import numpy as np
from pandas import read_csv
path = r"C:\pima-indians-diabetes.csv"
headernames = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age',
'class']
data = read_csv(path, names=headernames)
array = data.values
X = array[:,0:8]
127
Machine Learning with Python
Y = array[:,8]
data.shape
(768, 9)
data.head()
preg
Plas
Pres
skin
test
mass
pedi
age
class
0
6
148
72
35
0
33.6
0.627
50
1
1
1
85
66
29
0
26.6
0.351
31
0
2
8
183
64
0
0
23.3
0.672
32
1
3
1
89
66
23
94
28.1
0.167
21
0
4
0
137
40
35
168
43.1
2.288
33
1
patient_data = data.iloc[:, 3:5].values
import scipy.cluster.hierarchy as shc
plt.figure(figsize=(10, 7))
plt.title("Patient Dendograms")
dend = shc.dendrogram(shc.linkage(data, method='ward'))
128
Machine Learning with Python
from sklearn.cluster import AgglomerativeClustering
cluster = AgglomerativeClustering(n_clusters=4, affinity='euclidean',
linkage='ward')
cluster.fit_predict(patient_data)
plt.figure(figsize=(10, 7))
plt.scatter(patient_data[:,0], patient_data[:,1], c=cluster.labels_,
cmap='rainbow')